If False , subelements will not be processed when extracting info from elements. You can just call the corresponding method of the MzXML object. If the entity is a spectrum described in the offset index, it will be retrieved by immediately seeking to the starting position of the entry, otherwise falling back to parsing from the start of the file. Specify optional comma-separated pairs of Name,Value arguments. By default, mzxmlread reads all scans. 
| Uploader: | Domuro |
| Date Added: | 24 February 2010 |
| File Size: | 12.88 Mb |
| Operating Systems: | Windows NT/2000/XP/2003/2003/7/8/10 MacOS 10/X |
| Downloads: | 9676 |
| Price: | Free* [*Free Regsitration Required] |
The name of the compression method used before encoding the array into base The easiest solution is to just copy the dlls into the same directory as ReAdW. The Start scalar value must be less than End.

Once you have grabbed these dlls, you should avoid registering the XRawfile2. It is often coupled to chromatographic techniques such as gas- or liquid chromatography and has found widespread adoption in the fields of analytical chemistry and biochemistry where it can be used to identify and mzxnl small molecules and proteins proteomics. You can just call the corresponding method of the MzXML object.
Defines whether an index of byte offsets needs to be created for spectrum elements. Created using Sphinx 1.
Since you would still be missing zlib1. There are no planned further changes as of There is a viewer for ITA images. Go to the CVS repository.
Mass spectrometry data format
It can also link to CID and peptide probability information. Name must appear inside quotes. Here you will find the current draft of the mzXML ontology in Excel format. Select a Web Site Choose a web site to get translated content where available and see local events and offers.
Data from a single scan are converted to a human-readable dict. Input Arguments collapse all myFile — Input file character vector string.
It helps reduce memory usage at almost the same parsing speed. Element name or XPath-like expression. Random Access Minimal Parser. So MaxQuant seems to use some internal defaults for MS1 which cannot be modified by the user.
Read data from mzXML file - MATLAB mzxmlread
ANDI was initially developed for chromatography-MS data and therefore was not used in the proteomics gold rush where new formats based on XML were developed. Patrick Pedrioli was the primary original author; please see the references below for the often-cited Nature Biotech publication.
In this blog post we will elucidate the challenges of this endeavor and in the end provide several ways to generate MaxQuant-compatible mzXML. The program is based on the DAC library from Waters. Only local names separated with slashes are accepted.
If you do multiple iterfind calls on one file, you should create an MzXML object and use its iterfind method. Please see the massWolf page on the SPC Tools wiki for more information, including download instructions. The name of the XML attribute to use for lookup.
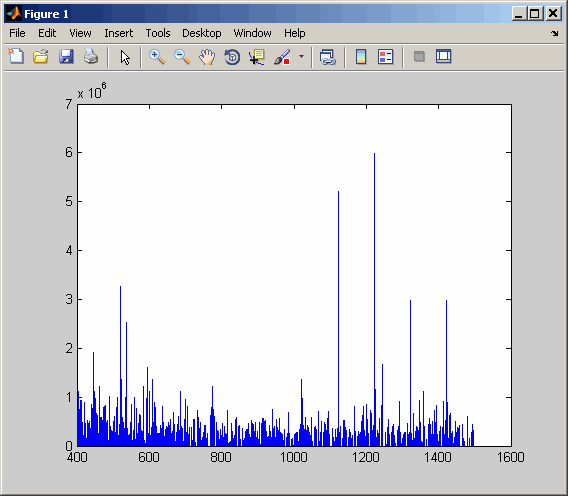
If you are using the 'Levels' name-value pair argument, then you cannot use 'TimeRange' or 'ScanIndices'. Choose a web site to get translated content where available and see local events and offers. The BPI can even be recomputed from the data.
Select a Web Site
A single, unifying data format for mass spectrometer output". ,zxml optional comma-separated pairs of Name,Value arguments. Since the mzXML needs to be indexed, you cannot simply edit an mzXML file manuallysince this will shift the byte offsets, and hence invalidate the index — and guess what happens then:

No comments:
Post a Comment